- Home
- LeishBASEedit Primer Design
- Plasmids
|
You are on LeishBASEedit!  |
|
Switch to... 
|
Switch to... 
|
|---|
What is LeishBASEedit?
LeishBASEedit is an online resource for using cytosine base editors in Leishmania and other kinetoplastids. Specifically, this website allows users to design primers suitable for cytosine base editing with the aim to introduce STOP codons within any given gene of interest (GOI). The STOP codon will be introduced by cytosine to thymine editing within the first 50% of the CDS. This will yield a functional null mutant (in most cases).
|
|
Contact |
|---|---|
|
17 Jun 2025: Our first genome-wide CRISPR base editing screen for L. mexicana is out together with our new database LeishBASEeditDB.net. Press the links to check out the paper Arias et al., BioRxiv 2025 and DATA. 27 Feb 2025: We just published v2 of LeishBASEedit in eLife: Herrmann May et al., eLife 2025. We show now how guide epxression constructs and libraries for base editing can be highly efficiently transfected and integrated using AsCas12a. 24 May 2023: LeishBASEedit is now published in eLife. Check out the final paper here: Engstler and Beneke, eLife 2023 8 Dec 2022: We are officially launching our new primer design site LeishBASEedit, which allows users to introduce STOP codons in their GOI using a cytosine base editor. You can read about it in our pre-print paper here: Beneke and Engstler, BioRxiv 2022 |
Please contact Dr. Tom Beneke (tobeneke@gmail.com or tom.beneke@uni-wuerzburg.de) at the University of Wuerzburg to request plasmids and for technical advice on primers, plasmids and cell lines. If you want to learn more about his research, please visit his lab website. |
|
Citation |
|
|
Please cite both our publications when using the LeishBASEedit website and method in any given organism: Engstler and Beneke, eLife 2023 and Herrmann May et al., eLife 2025. |
|
Acknowledgements Special thanks go to Prof. Markus Engstler at the University of Wuerzburg for supporting the development of the LeishBASEedit toolbox. Tom Beneke is funded by an EMBO (ALTF 727-2021) and Marie Curie (ID 101064428) Postdoctoral Fellowship.
Website designed and maintained by Tom Beneke (tobeneke@gmail.com or tom.beneke@uni-wuerzburg.de). Last updated on 17th June 2025. |
LeishBASEedit Primer Design v2
LeishBASEedit is a primer design tool to introduce STOP codons into any given gene of interest (based on TriTrypDB release 59). Our toolbox, described in Engstler and Beneke, eLife 2023 and Herrmann May et al., eLife 2025, uses the hyBE4max cytosine base editor from the Li lab (Zhang et al., Nature Cell Biology 2020). This optimised cytosine base editor, has an expanded cytosine editing window within the single-guide RNA target sequence (editing window is between nucleotide position 4 and 12). To use the hyBE4max CBE in Leishmania, we have generated multiple CBE expression plasmids. Guides for base editing can be cloned into plasmids containing a ribosomal promoter or a modified T7 RNAP promoter (T7 T10 GG variant) for sgRNA expression. You can view these under the plasmid tab. As shown below, these guide expression constructs can be highly efficiently delivered and integrated into the Leishmania genome using AsCas12a (for a detailed method description see Herrmann May et al., eLife 2025).
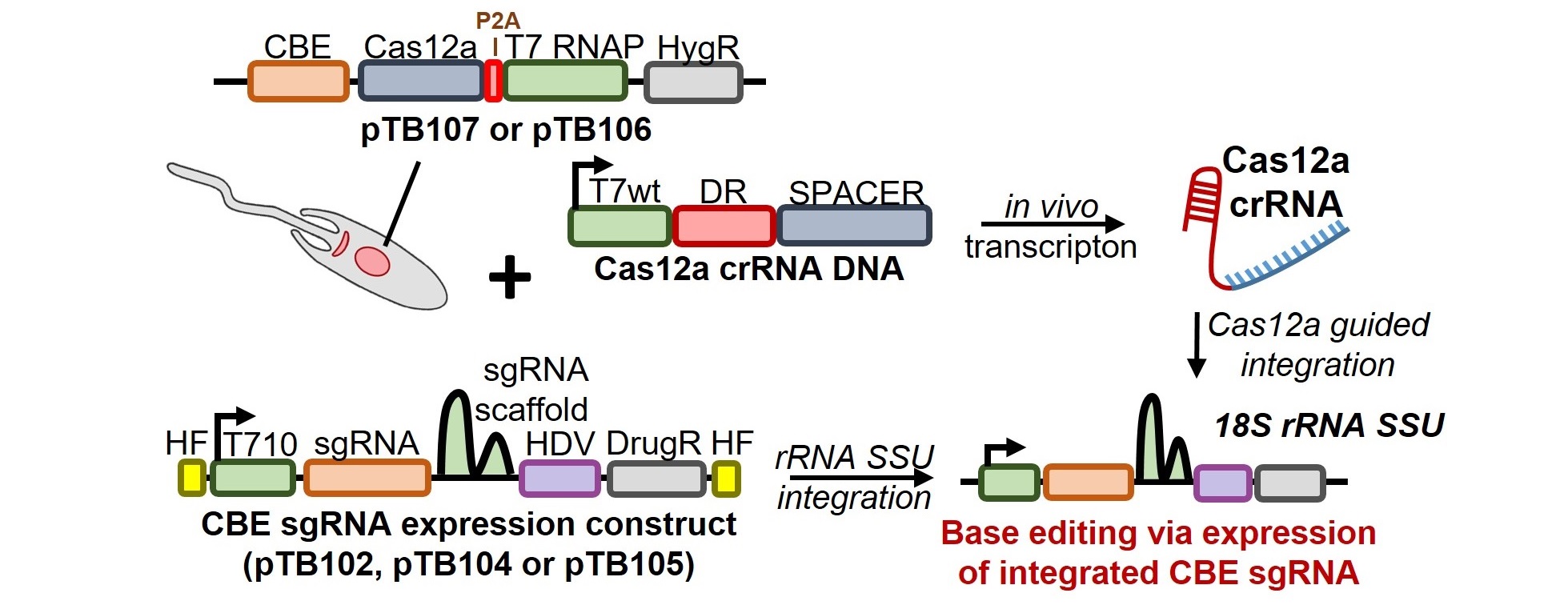
In the search window below, users can design oligos that are needed to clone a single-guide RNA into guide expression plasmids. Guide RNAs are designed through a CRISPR-CBEI pipeline (Yu et al., mSystems 2020). Below is a detailed explanation of how this primer design pipeline works and how to interpret the primer design output.
At present, there are a total of 11,457,418 primer sequences available for 64 different species (based on TriTrypDB release 59). Please cite our website and publication (Engstler and Beneke, eLife 2023 and Herrmann May et al., eLife 2025) so that we can continue this resource for the kinetoplastid community.
A protocol for cloning the guides into guide expression plasmids can be downloaded here: |
A protocol for using Cas12a for stable integration of CBE guide expression constructs into Leishmania parasites can be downloaded here: |
A protocol for transfecting plasmids into Leishmania parasites can be downloaded here: |
Please use the form below to retrieve single-guide RNAs and primer sequences for your gene of interest. The guide ranking is based on our updated ranking tool (LeishBASEedit v2: Herrmann May et al., eLife 2025). Please use the downloadable '.csv' files for larger querries.
Design Primer Sequences
|
Just enter your GeneIDs below You can export your designed primers as a '.txt' file on the search result page. |
|---|
You can download a '.csv' file with a complete list of all LeishBASEedit primer designs for any of the following organisms (annotations based on TriTrypDB release 59):
How are LeishBASEedit primers and guides designed?
Cytosine base editors are comprised of a cytidine deaminase domain (e.g. APOBEC-1) fused to an impaired form of Cas9 (e.g. D10A nickase) and to two monomers of uracil glycosylase inhibitors (UGI). Cytosine base editors do not require donor DNA or the introduction of double-strand breaks for gene editing. The deaminase domain facilitates a deamination, which transforms cytidine to uridine. Uridine is then repaired to thymidine during DNA replication or other DNA repair processes.
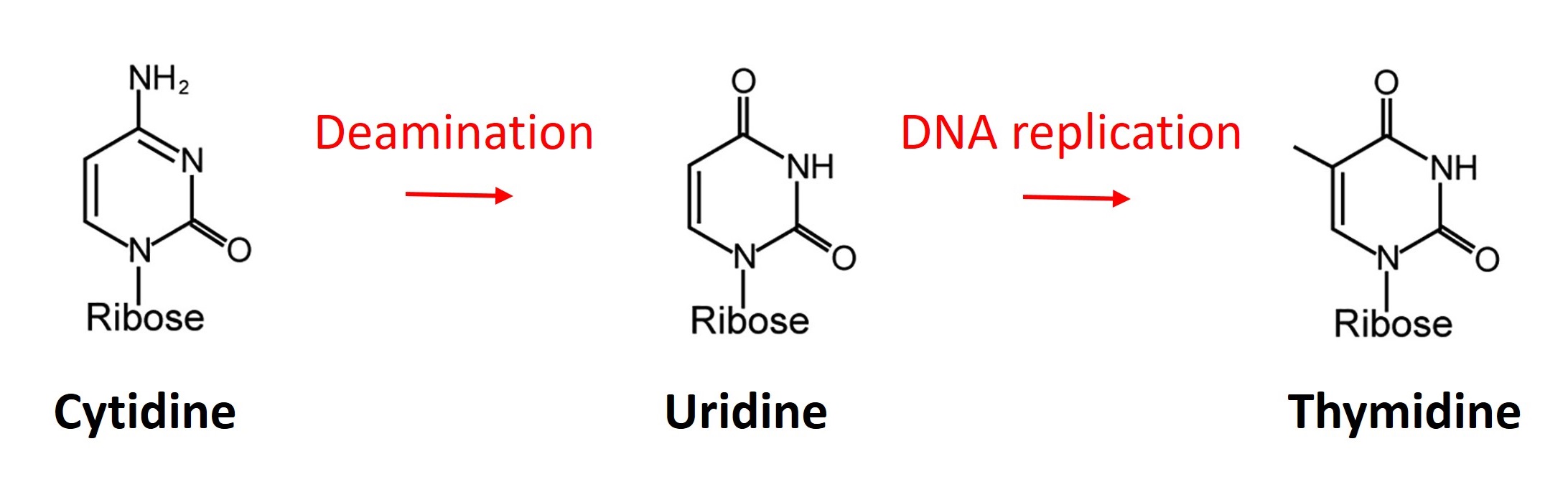
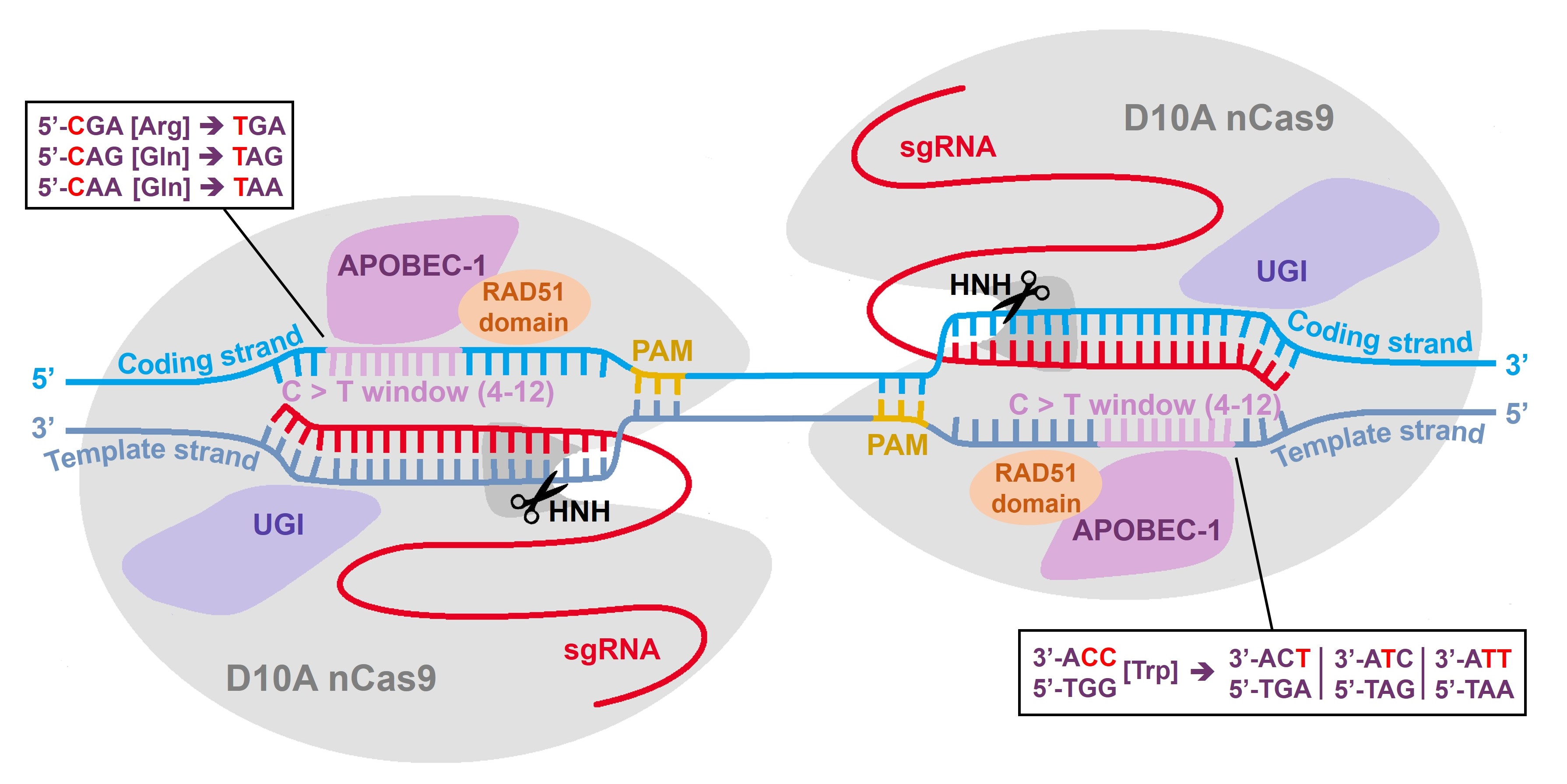
Cytosine base editors have specific editing windows within the single-guide RNA sequence, in-which cytidine to thymidine editing is efficient. In case of the hyBE4max editor from Zhang et al., Nature Cell Biology 2020, the editing window is at position 4-12 of the guide RNA target sequence (see figure below). There are four codons that result in a STOP codon, when cytidine is edited to thymidine (see figure below). LeishBASEedit designs single-guide RNAs that catalyse the editing of cytidine to thymidine in this 4-12 editing window and filters out only guides that target one of these four codons to transform them into a STOP codon. Guides are then filtered for targeting only the first 50% of the CDS. The assumption is that (in most cases) a functional mutation can be achieved by introducing a STOP codon within the first half of any given gene. In addition, guide ranking is also affected by the position of the STOP codon within the editing window (e.g. 4-8 has higher activity than 8-12) and by the number of possible STOP codons that could be introduced. Lastly, guides are ranked to have a low cytidine to STOP codon ratio. This allows to prioritize CBE sgRNAs that ensure STOP codon generation and eliminates competition between deleterious and non-deleterious mutations. You can find more infos about guide scoring in our recent toolbox release (LeishBASEedit v2: Herrmann May et al., eLife 2025).
Shown below is a schematic of the hyBE4max base editor and how it can be used to introduce STOP codons. For a detailed description please read our publications: Engstler and Beneke, eLife 2023 and (LeishBASEedit v2: Herrmann May et al., eLife 2025).
Understanding the LeishBASEedit primer output
Upon submission of a gene ID into the search window above, LeishBASEedit v2 will retrieve detailed information for all possible single-guide RNAs for the queried gene, as well as two primers that can be annealed and then cloned into a sgRNA expression plasmid (you can download the cloning protocol above). The figure below shows a primer design example for one of the guides in the PF16 gene (LmxM.20.1400). The left panel shows the cloning scheme that can be followed with the retrieved primers from LeishBASEedit (top left panel) and the guide scoring/ranking scheme as described in Herrmann May et al., eLife 2025(bottom left panel). Primers are provided for cloning into rRNA and T7 T10 GG promoter-driven sgRNA expression vectors. The panel on the right explains how to interpret the information provided by LeishBASEedit for each guide target sequence (following CBEI definitions: Yu et al., mSystems 2020).
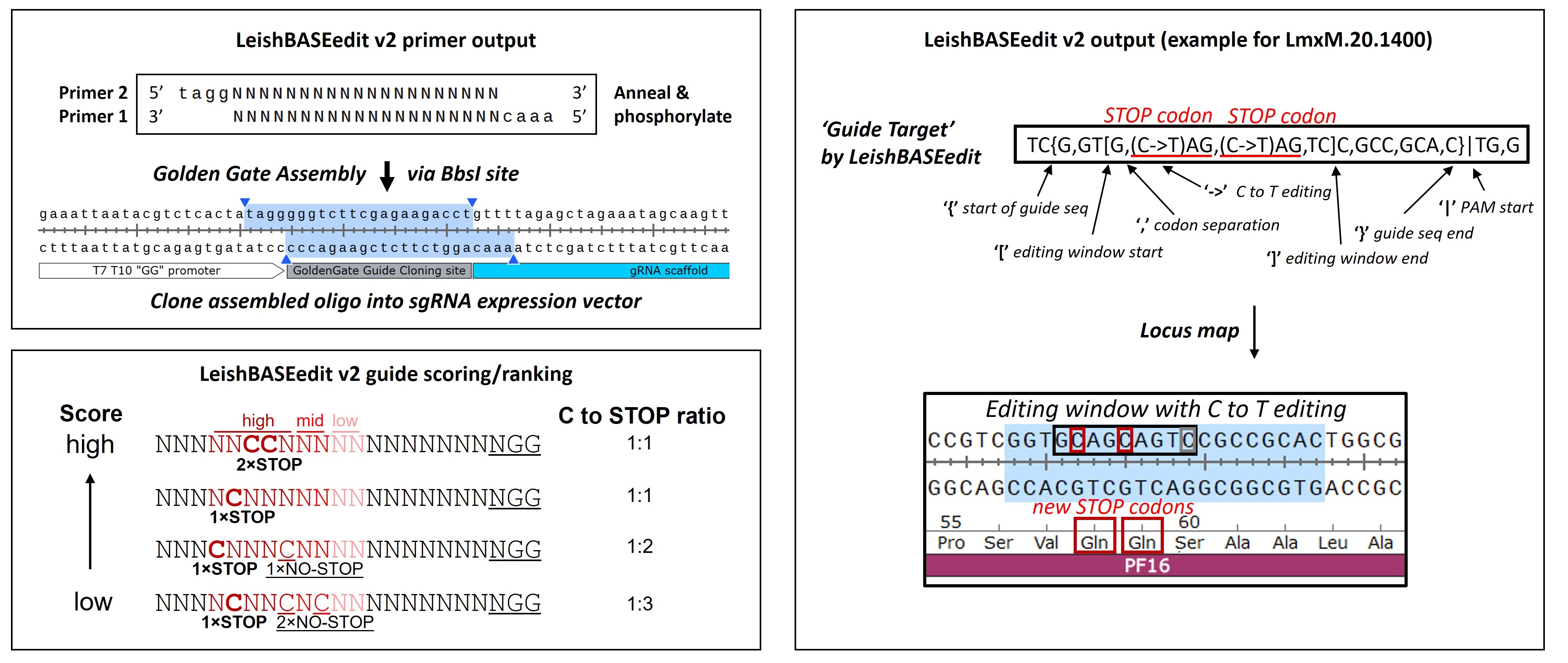
Other LeishBASEedit primer output parameters
In addition to the 'Guide Target' output and the provided primers 1 and 2, LeishBASEedit prints out information for each guide designed for the gene of interest. An explanation for each of those parameters is provided in the table below.
| LeishBASEedit output | Description |
|---|---|
| Gene ID | Gene ID querry by user |
| Guide ID v2 | Unique guide ID assigned by LeishBASEedit v2 |
| Strand | DNA strand where C to T editing takes place |
| CDS Position [%] | Percentage of the CDS that would remain if STOP codon is introduced |
| Guide Target | Guide target schematic as explained in figure above (panel on the right) |
| Guide Sequence | 20 nt single-guide RNA sequence |
| Guide Coordinates | Coordinates of single-guide RNA sequence (start from 'ATG' of given gene) |
| Stop Codon Coordinates | Coordinates of newly introduced STOP codons (start from 'ATG' of given gene) |
| Editing Window | Coordinates of editing window (start from 'ATG' of given gene) |
| PAM Sequence | PAM sequence of given single-guide RNA |
| PAM Position | Coordinates of PAM sequence (start from 'ATG' of given gene) |
| Edited Sequence | Proceeding nucleotide and edited nucleotide of new STOP codon |
| Perfect match count | Count of the perfect match of guide target sequence and its adjacent PAM site in the target genome |
| Number of Cytosines | Number of cytosines within the 4-12nt editing window |
| Number of possible STOP codons | Number of possible STOP codons when C is converted to T within the 4-12nt editing window |
| Ratio C to STOP | Number of cytosines divided by number of possible STOP codons (both within the 4-12nt editing window) |
| Edit Window 4-8 | Checks whether the C to T edit is within the 4-8 editing window |
| Edit Window 4-10 | Checks whether the C to T edit is within the 4-10 editing window |
| C score | Guide score (see Herrmann May et al., eLife 2025 for more info) |
| Primer 1 (guide reverse) | First of the two primers needed to clone the guide into sgRNA expression vectors (sequence needs no further modification for primer order) |
| Primer 2 (rRNAP guide forward) | Second of the two primers needed to clone the guide into rRNA promoter sgRNA expression vectors (sequence needs no further modification for primer order) |
| Primer 2 (T7 T10 GG guide forward) | First of the two primers needed to clone the guide into T7 T10 GG promoter sgRNA expression vectors (sequence needs no further modification for primer order) |
Website designed and maintained by Tom Beneke (tobeneke@gmail.com or tom.beneke@uni-wuerzburg.de). Last updated on 17th June 2025.
Plasmids for gene editing
Plasmids below can be used for base editing in Leishmania. A detailed description of each plasmid can be found in Engstler and Beneke, eLife 2023 and Herrmann May et al., eLife 2025. Please contact Tom Beneke (tobeneke@gmail.com or tom.beneke@uni-wuerzburg.de) for plasmid requests, as well as for technical advice on primers, plasmids and cell lines.
| Plasmid Name | Promoter | Plasmid purpose | Resistance Marker | Size | GeneBank file |
|---|---|---|---|---|---|
pLdCH-hyBE4max |
L. donovani rRNA promoter |
Co-expression of sgRNA and hyBE4max | Hygromycin | 11,160 bp | |
pLdCH-hyBE4max-LmajDBD |
L. donovani rRNA promoter |
Co-expression of sgRNA and hyBE4max with Rad51 ssDNA binding domain (DBD) from L. major Friedlin | Hygromycin | 11,271 bp | |
pTB007-hyBE4max |
Intergenic driven |
Co-expression of hyBE4max and T7 RNAP | Hygromycin | 17,171 bp | |
pTB106 |
Intergenic driven |
Co-expression of hyBE4max-LmajDBD, T7 RNAP and AsCas12a ultra (M537R/F870L mutation) | Hygromycin | 21,608 bp | |
pTB107 |
Intergenic driven |
Co-expression of hyBE4max, T7 RNAP and AsCas12a ultra (M537R/F870L mutation) | Hygromycin | 21,498 bp | |
pTB102 |
T7 T10 GG promoter variant |
Expression of sgRNA; plasmid without homology arms for integration | Puromycin | 3,849 bp | |
pTB104 |
T7 T10 GG promoter variant |
Expression of sgRNA; plasmid with homology arms for integration into NeoR gene | Puromycin | 4,557 bp | |
pTB105 |
T7 T10 GG promoter variant |
Expression of sgRNA; plasmid with homology arms for integration into 18S rRNA SSU locus | Puromycin | 4,491 bp |
Technical notes:
Below users can download protocols for guide cloning, Cas12a-mediated integration of expression constructs and transfection of DNA constructs into Leishmania parasites. Please cite Engstler and Beneke, eLife 2023 and Herrmann May et al., eLife 2025 when using these protocol.
A protocol for cloning the guides into guide expression plasmids can be downloaded here: |
A protocol for using Cas12a for stable integration of CBE guide expression constructs into Leishmania parasites can be downloaded here: |
A protocol for transfecting plasmids into Leishmania parasites can be downloaded here: |
Website designed and maintained by Tom Beneke (tobeneke@gmail.com or tom.beneke@uni-wuerzburg.de). Last updated on 17th June 2025.
 What is new on LeishBASEedit?
What is new on LeishBASEedit?